Software >Clustering Using Betweenness Centrality
Description | Pros and Cons | Applications | Details | Usage Hints | References | Acknowledgments| Description |
This clustering algorithm uses Brandes' algorithm to calculate the 'betweenness centrality' for vertices. Subsequently, the betweenness centrality of the edges within a network is calculated and the edge with the maximum betweenness centrality score is removed. This is repeated until a user given threshold value is higher than the betweenness centrality scores of all the edges or until the network is split into a certain number of subnetworks.
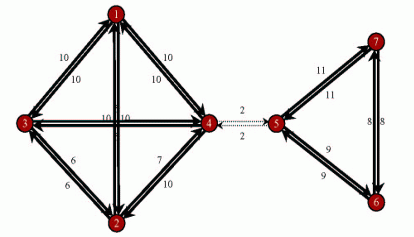
A network divided
into 2 subnetworks by removal of the weak links (dashed)
The first definition
of betweenness centrality came from Anthonisse and Freeman:
The computation of betweenness centrality is computationally
very expensive. Networks of up to several hundred nodes can be analyzed,
but the original algorithm does not scale to
networks with thousands of nodes. In 2001, Ulrik
Brandes proposed a more efficient algorithm
for betweenness that exploits the extreme sparseness of typical
networks to reduce running time from O(n3) to O(n2 + nm) and memory
consumption from O(n2) to O(n+m) [3] (n
is the number of nodes and m is the number of edges). Moreover, other
shortest-path based indices, like closeness or radiality, can be
computed simultaneously within the same bounds.
Given the betweenness centrality values for all nodes, the betweenness centrality scores of edges can be calculated. Edges with the maximum score are assumed to be important for the graph to stay interconnected. These high scoring edges are the 'weak ties' that interconnect clusters of nodes. Removing them leads to unconnected clusters of nodes.
A breadth-first search algorithm for unweighted graphs (and Dijkstra's algorithm for weighted graphs) was implemented to calculate the number of shortest paths between any two nodes and to get the set of all predecessor nodes. Both pieces of information are required to calculate the betweenness centrality according to Brandes' formula.
The breadth-first search explores a graph G=(V,E) across the breadth of the frontier between discovered and undiscovered nodes. A node is examined more, the farther its distance from the starting node s. That is, the algorithm discovers all nodes at distance k from the starting node s before discovering any nodes at distance k+1. As a result, one gets the shortest paths to each node v of V that is reachable from s. A shortest path from s to v is the path which consists of the lowest number of edges. To describe the current state of the breadth-first search every node can be colored white, gray or black. At the beginning all nodes are white. When a node gets visited for the first time it is colored non-white. That is, all gray and black nodes have been visited at least once. An already visited node gets the color black if all its child nodes have already been visited (which means that it is colored non-white). On the other hand, an already visited node gets the color gray if it has at least one child node which is not visited yet (which means that it is white). Therefore, the gray nodes describe the frontier between the visited and not yet visited nodes.
The pseudo code for the relaxation procedure for a single edge is as follows:
define relax(Node u, Node v, int weight)
if d(u) + weight < d(v) then
d(v) := d(u) + weight
pi(v) := u
The pseudo code for Dijkstra's Algorithm is as follows:
define dijkstra(Graph g, Node s)
initializeSingleSource(g, s)
S := {}
Q := getNodes(g)
while not empty(Q)
Node u := extractMin( Q )
AddNode( S, u )
for each node v in neighbors( u )
relax( u, v, weight(u,v) )
define initializeSingleSource(Graph g, Node s)
for each node v in getNodes(g)
d(v) = INFINITY
pi(v) = NIL
d(s) = 0
pi(s) = NIL
| Pros & Cons |
Applicable to network data only. The algorithm
does not scale to more than 5000 nodes.
| Applications |
The algorithm has been successfully applied to identify clusters of co-author networks, interrelated genes, etc.
| Details |
There are two versions of this clustering algorithm - for weighted and unweighted graphs. The former uses a breadth-first search algorithm and integer data structure to store 0 and 1 for the graph. The other one will work on both unweighted graphs and weighted graphs, but is inefficient for unweighted graphs as it uses Dijkstra's algorithm to find the shortest path and floating point data structures to store the graph. Although the results of both algorithms are identical there are considerable difference in running time for large scale networks.
| Usage Hints |
1. Download and uncompress
the zipped files on your computer using winzip or other utility.
2. It should make a directory named src in the directory you have
downloaded it in.
3. There will be three files: BCclustering.java,
input.txt, output.txt. The two .txt files are input and
output sample files respectively and the .java file is the original
source file.
4. To compile it, issue the command javac BCclustering.java from
the same directory. It will make several class files upon successful compilation.
5. To run it, issue the command
java BCclustering input.txt output.txt. It will read input from the input
file and recreate the output file.
6. Enter zero to see the Centrality Scores of all the edges or enter
a threshold value when asked.
7. After each iteration, the current graph will be stored in the
file named out_tmp.txt just in case the user aborts the program or it crashes. When reinvoking this file it should be copied with some
other name and given as the input filename to the program. It can also
be used to see the current network status at any point of program execution.
Currently, the program is
tested by compiling with the JDK version-1.4.0.00. It stores the result
in the form of an adjacency matrix of the remaining part of the graph and also takes
the input in the same format. Visone can be used to visualize the input and output
networks and can be downloaded for free from www.visone.de.
Download visone, uncompress and run the program and then go to File->import->matrix
and select the file to visualize.
| References |
- Anthonisse, J.M.(1971) The rush in a directed graph. Technical Report BN9/71, Stichting Mahtematisch Centrum, Amsterdam.
- Freeman, L.C.(1977) A set of measuring centrality based on betweenness. Sociometry 40, 35-41.
- Ulrik Brandes (2001) A
Faster Algorithm for Betweenness Centrality. Journal of Mathematical
Sociology 25(2):163-177.
| Acknowledgments |
The algorithm was implemented and documented by Vivek Agrawal (vivekagr@iitk.ac.in).
![]()
Information
Visualization Cyberinfrastructure @ SLIS,
Indiana University
Last Modified January
14, 2004